Last 26th July 2023 I successfully defended my PhD Thesis entitled: “PhD Thesis: Implementation of high-resolution MRSI methods in a pre-clinical scanner, and optimization for brain longitudinal studies of therapy response in mice glioma model.”
Abstract
Magnetic resonance spectroscopy and magnetic resonance spectroscopic imaging (MRS/MRSI) are non-invasive diagnostic techniques that use a strong magnetic field and radio waves to examine the chemical composition of living tissue. Working on the same principles as Magnetic Resonance Imaging (MRI), instead of producing images, MRS generates a spectrum of signals that can be used to identify the type and amount of molecules present in a tissue. The utility of MRS and MRSI has already been established in many studies, providing useful information about the chemical makeup of different regions of the brain, and allowing diagnosis of conditions such as Alzheimer’s disease, multiple sclerosis, and brain Glioblastoma (GB) tumors.
Preclinical glioblastoma studies looking forward to improving therapeutic outcomes are necessary since clinical GB has no current cure. These studies can greatly benefit from improved spatial resolution and homogeneity of the acquired MRSI grids. Hence, we can work towards improved acquisition schemes enhancing the quality of acquired data using MRS and MRSI. There exists a methodological consensus among spectroscopy experts where the Localized Adiabatic Spin Echo Refocused (semiLASER) data acquisition strategy has been ranked as the most likely localization technique to improve (pre) clinical MRS. SemiLASER uses adiabatic pulses to selectively excite and refocus the signal from a localized volume of interest in the brain. This results in a higher signal-to-noise ratio (SNR) and better spatial resolution compared to conventional data acquisition sequences.
Partial volume effects can occur in MRSI when the voxel (a 3D volume of interest) being measured contains a mixture of different neighbouring tissue types or compartments, such as grey and white matter or cerebrospinal fluid. This can lead to inaccurate quantification of metabolites, as the signal from one tissue can mix with the signal from another and affect overall pattern recorded. SemiLASER is designed to minimize partial volume effects by using adiabatic pulses to selectively excite and refocus the signal from a small region of interest within the voxel. This allows for more accurate quantification of metabolites within the region of interest, while reducing the contamination of the signal by other tissue types. In addition, semiLASER also employs an outer-volume suppression (OVS) technique to further reduce contamination from outside the region of interest. This involves using additional adiabatic pulses to selectively saturate the signal from outside the volume of interest, so that it does not contribute to the MRSI signal. Overall, the combination of selective excitation and OVS in semiLASER can help improve the accuracy of MRSI measurements and reduce partial volume effects.
Although, the clinical utility of semiLASER has been acknowledged, the pre-clinical use and implementation of semiLASER still remains a less explored area. Our group has a long record of using MRSI in therapy response monitoring of a murine model glioblastoma (the GL261 cell line) using a commercially available MRSI acquisition sequence. In our efforts towards bridging the barriers between pre-clinical and clinical research, we have implemented the clinically verified semiLASER sequence on a pre-clinical 7T Bruker Biospec USR scanner running the ParaVision 5.1 software package, which provides a graphical user interface for sequence programming and data acquisition. The single and multi-voxel semiLASER sequences were implemented with the idea that the developments generated during this PhD project will be replicable by other interested users.
The implemented SV-semiLASER and MRSI-semiLASER sequences for preclinical acquisitions were optimised to perform high resolution MRSI of living mouse brain. For this, sequences were duly verified and tested first in phantoms and later in-vivo, in wild-type (wt) and tumor bearing (GL261) mice. To do so, the Bruker pulse sequence implementation was first studied in detail to become familiar with the Bruker programming environment and a test sequence PRESS_Slice to localize the slice dimension was developed by modifying the Bruker stock PRESS sequence for single voxel localization. After careful evaluation of test sequence results, the semiLASER single and multi-voxel sequences were also implemented using a similar strategy.
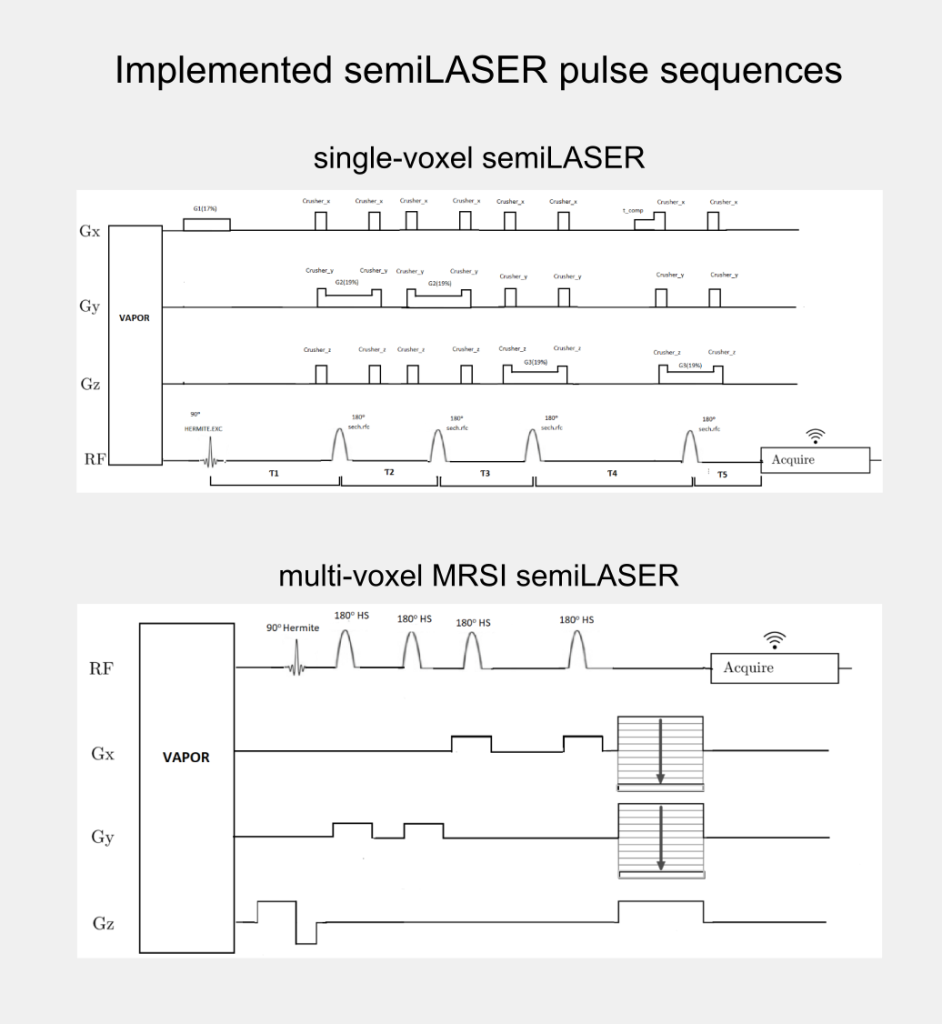
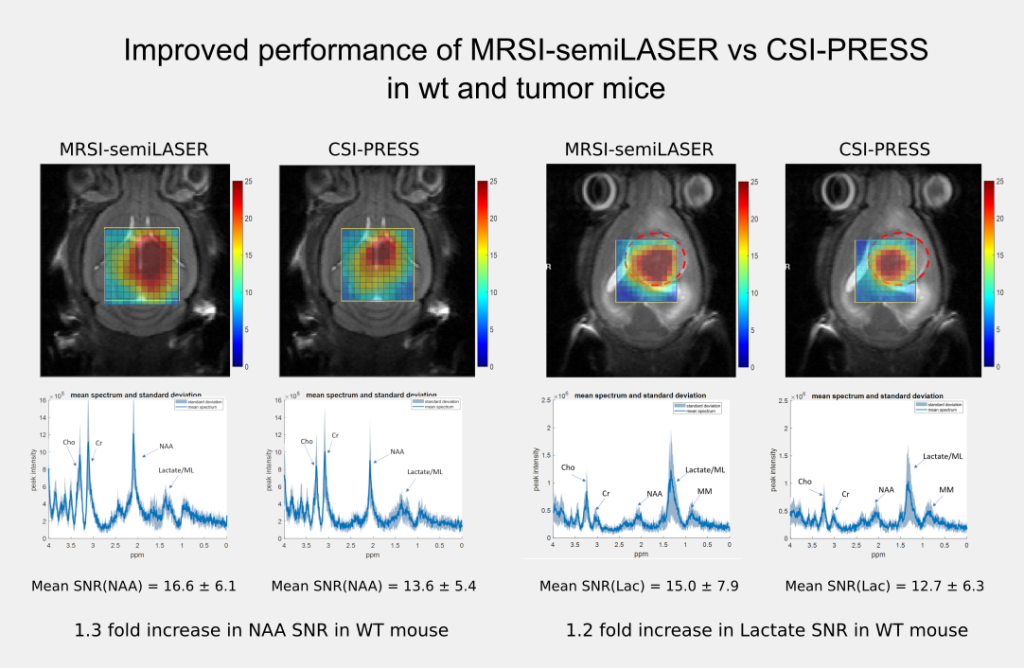
The implemented SV-semiLASER sequence provided a ca. 1.4-fold improvement in SNR in phantoms and ca. 1.3-fold improvement in SNR for in-vivo subjects, in comparison to the stock Bruker PRESS (single volume acquisition) sequence. The MRSI-semiLASER sequence resulted in a ca. 1.3-fold improvement of SNR in phantoms and in-vivo subjects compared to the stock Bruker CSI-PRESS sequence. Combined with phase encoding strategies and volume reduction methods, higher spatial resolution and SNR was achieved for the implemented MRSI-semiLASER. The quantification analysis of the results was done using MATLAB based post-processing tools specially designed to process Bruker datasets and solutions for a faster post processing pipeline were proposed. The single voxel MRSI-semiLASER sequences were further simulated using NMRSCOPE-B virtual simulator, a jMRUI plug-in which positively correlated with the experimental results. Preliminary nosological images obtained using MRSI-semiLASER datasets and the SpectraClassifier tool previously developed in our group, and trained with GL261 tumors using already available CSI-PRESS data, suggested those classifiers could be robust enough to recognize the tumor region acquired with the semi-LASER sequence. Still, classifiers may require retraining for the evaluation of response to therapy, which is an ongoing project within the group.
The thesis dissertation can be downloaded in PDF format using the link below or from the official TXD and Teseo repositories (currently in progress):
Acknowledgments
I would like to thank the financial support by the European Comission Marie Curie Initial Training Networks (ITN, call H2020-MSCA-ITN-2018, grant 813120 to project INSPiRE-MED); by the Ministry of Science and Insdustry (MCIN/AEI/10.13039/501100011033) (APC); and by Centro de Investigación Biomédica en Red—Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN http://www.ciber-bbn.es/en, CB06/01/0010), an initiative of the Instituto de Salud Carlos III (Spain) co-funded by the European Regional Development Fund (ERDF). I was recipient of a Marie Skłodowska-Curie early-stage researcher fellowship of the INSPiRE-MED project (Grant agreement ID: 813120)